Signature¶
The signature is an array that assigns a probability to a single nucleotide mutation taking into account its context [1]. It represents the chance of a certain mutation to occur within a context.
Check the different options for the signature in the configuration file. In short, you can choose between not using any signature, using your own signature or computing the signature from the mutations file. Additionally, signatures can be grouped into different categories (such as the sample).
The signature array is computed by counting, for each Single Nucleotide Polymorphism, the reference and alternated triplets.
Note
OncodriveFML also uses the MNP mutations to compute the
signature, by treating them as a set of separate SNPs.
You can enable or disable this behaviour with the include_mnp option in the
configuration file.
The counts are then divided by the total number of counts
to generate a frequency of triplets. For a mutation 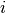 the frequency is
the frequency is
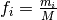 where
where 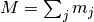 , and
, and
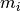 represent the number of times that the mutation
represent the number of times that the mutation
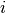 with its context [1] has been observed.
with its context [1] has been observed.
Optionally, the signature can be corrected taking into
account the frequency of trinucleotides in the
reference genome.
OncodriveFML introduces this feature because the
distribution of triplets is not expected to be constant.
When using the command line interface, OncodriveFML
does this correction automatically according to
the value passed in the flag --sequencing
(you can list all the options using the help).
Reasoning behind the correction¶
Let’s first take the conditional probability of a mutation (with contectx [1])
to occur given the number of those triplets in the region:
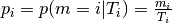 .
.
Then, the normalized frequency of the mutation 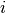 is:
is:
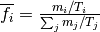 .
.
The results can be adapted in case our inputs are not absolute values but relative frequencies.
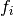 is the frequency of mutations and
is the frequency of mutations and 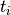 the frequency of nucleotides:
the frequency of nucleotides:
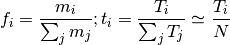
(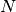 is the number of nucleotides,
is the number of nucleotides, 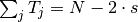 , where
, where  is the number of segments)
is the number of segments)
Then:
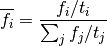
Proof:
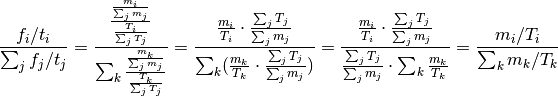
| [1] | (1, 2, 3) The context is formed by the previous and posterior nucleotides. |